Magazine

CAD4TB, a crucial diagnosis aid
CAD4TB, acronym for «computer-aided detection for tuberculosis», is here to help healthcare professionals around the globe in the diagnosis of active tuberculosis, a disease that still has huge incidence and mortality rates, particularly in poor countries. How does the CAD4TB algorithm work? The CAD4TB algorithm is a code that works with high-quality DICOM posterior-anterior chest X-ray images form patients above 16 years old. The algorithm detects the shape of the lungs and evaluates their texture. It superimposes a «heat map» over lung areas, where blue indicates normal areas and green/yellow/orange/red colours indicate areas with increasing probability of tuberculosis-like abnormalities, i.e. areas with abnormal textures potentially caused by active tuberculosis. Besides providing this heat map, the algorithm generates a score that indicates the probability of active tuberculosis on a scale from 0 (low probability) to 100 (high probability). CAD4TB algorithm: low score. Left: X-ray image; right: CAD4TB algorithm heat map. CAD4TB algorithm: high score. Left: X-ray image; right: CAD4TB algorithm heat map. Is a high CAD4TB score enough to diagnose active tuberculosis? Healthcare professionals set a certain threshold for the score; if a patient’s score is higher than that value, a sputum test has to be performed in order to confirm the diagnosis. The score does not confirm the disease per se, but allows patients to be referred to a definite diagnosis test only when there is a suspicious high score, which helps bringing down the diagnosis costs, particularly when performed on a big population. Why was this algorithm developed? The developers of the CAD4TB algorithm aimed at providing a quick and easy solution to detect tuberculosis. This algorithm was intended to be used, right from its conception, in low-income, developing countries—those where the prevalence of tuberculosis is higher. The World Health Organization (WHO) had already recommended X rays as a systematic method for tuberculosis screening, given their low operating costs and quick results; nonetheless, despite these cost/speed benefits, local expertise is still needed to interpret the actual X-ray images. This is where the CAD4TB algorithm plays a key role: it automatically flags potential positive cases of active tuberculosis, and those suspicious cases can be confirmed remotely by experts based anywhere in the globe (thanks to a cloud-based technology named CAD4TBcloud). Who developed this algorithm? The CAD4TB software was developed by the Diagnostic Image Analysis Group (a group that belongs to the Departments of Radiology and Nuclear Medicine, Pathology and Ophthalmology of the Radboud University Medical Center, The Netherlands) and Delft Imaging Systems (a company specialized in the development of diagnostic imaging devices based in Veenendaal, The Netherlands). Exclusive distribution rights are licensed to Delft Imaging Systems. Further development of this algorithm is now performed by Thirona, a spin-off company from the Diagnostic Image Analysis Group. For its development, this system used machine learning to learn from images interpreted manually and also received input from leading lung specialists across the globe. Tuberculosis Tuberculosis is still a high-burden disease. According to the latest report of WHO on tuberculosis, it infected around 10 million people in 2017 and is one of the top 10 causes of death.
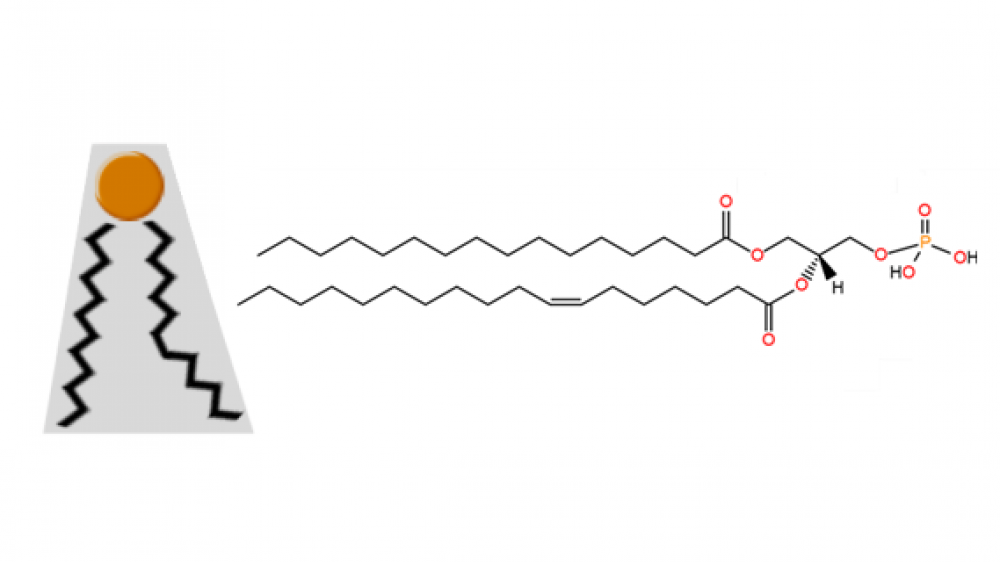
Phosphatidic acid structure
The structure of phosphatidic acid is very simple; indeed, it is the simplest diacyl glycerophospholipid. It consists of a glycerol backbone linked to one phosphate group and two acyl chains. This lipid accounts for approximately 1-4% of the total lipids present in the cells of any living organism. An example of a molecule of this phospholipid is shown in Figure 1 below. In this case, it contains the acyl chains palmitoyl (16:0) and oleoyl (18:1). The most common fatty acids found in naturally occurring phosphatidic acid molecules are palmitic, oleic, stearic (18:0), linoleic (18:2) and arachidonic (20:4) acids. Figure 1. Organic structure of one molecule of phosphatidic acid; a) acyl chains; b) glycerol backbone; c) phosphomonoester group; the carbon molecules of glycerol are numbered in blue. The small size of its phosphate head group, compared to the size of its base, confers it a particular cone shape (Figure 2 below) that is very important in lipid bilayers. This shape is key to allow the formation of negative curvatures of lipid bilayers that is critical to the fusion and fission of portions of membranes or entire organelles (e.g. plasma membrane or Golgi apparatus), as well as to various biochemical processes happening near cell membranes (e.g. recruitment of signaling proteins). Figure 2. Cone shape of a molecule of phosphatidic acid; the small head group represented in orange; the black zigzag lines represent acyl chains. The free phosphate group provides phosphatidic acid with a negatively charged head group. These anionic lipids are known to recruit and dock positively charged molecules. Plus, its phosphomonoester group can carry one or two negative charges; inside cells, the physiological pH of different intracellular situations provides the conditions to make phosphatidic acid carry one or two negative charges, which consists on a variable that can be used by certain cell environments to start or end PA-dependent signaling cascades. Further reading: Ferraz-Nogueira JP, Díez-Guerra FJ, Llopis J. Visualization of phosphatidic acid fluctuations in the plasma membrane of living cells. PLoS One. 2014 Jul 15;9(7):e102526. Tanguy E, Kassas N, Vitale N. Protein⁻Phospholipid Interaction Motifs: A Focus on Phosphatidic Acid. Biomolecules. 2018 Apr 23;8(2).
